Characteristic Gene Selection and Tumor Classification by RLSDSPCA
Abstract: Characteristic gene selection and tumor classification of gene expression data play major roles in genomic research. Due to the characteristics of a small sample size and high dimensionality of gene expression data, it is a common practice to perform dimensionality reduction prior to the use of machine learning-based methods to analyze the expression data. In this context, classical principal component analysis (PCA) and its improved versions (e.g., sparse principal component analysis (SPCA) and graph regularized PCA) have been widely used. Recently, methods based on supervised discriminative sparse PCA (SDSPCA) have been developed to improve the performance of data dimensionality reduction. However, such methods still have limitations: most of them have not taken into consideration the improvement of robustness to outliers and noise, label information, sparsity, as well as capturing intrinsic geometrical structures in one objective function. To address this drawback, in this study we propose a novel PCA-based method, known as robust Laplacian supervised discriminative sparse PCA, termed RLSDSPCA, which enforces the L2,1 norm on the error function and incorporates the graph Laplacian into supervised discriminative sparse PCA. To evaluate the efficacy of the proposed RLSDSPCA, we applied it to the problems of characteristic gene selection and tumor classification problems using gene expression data.
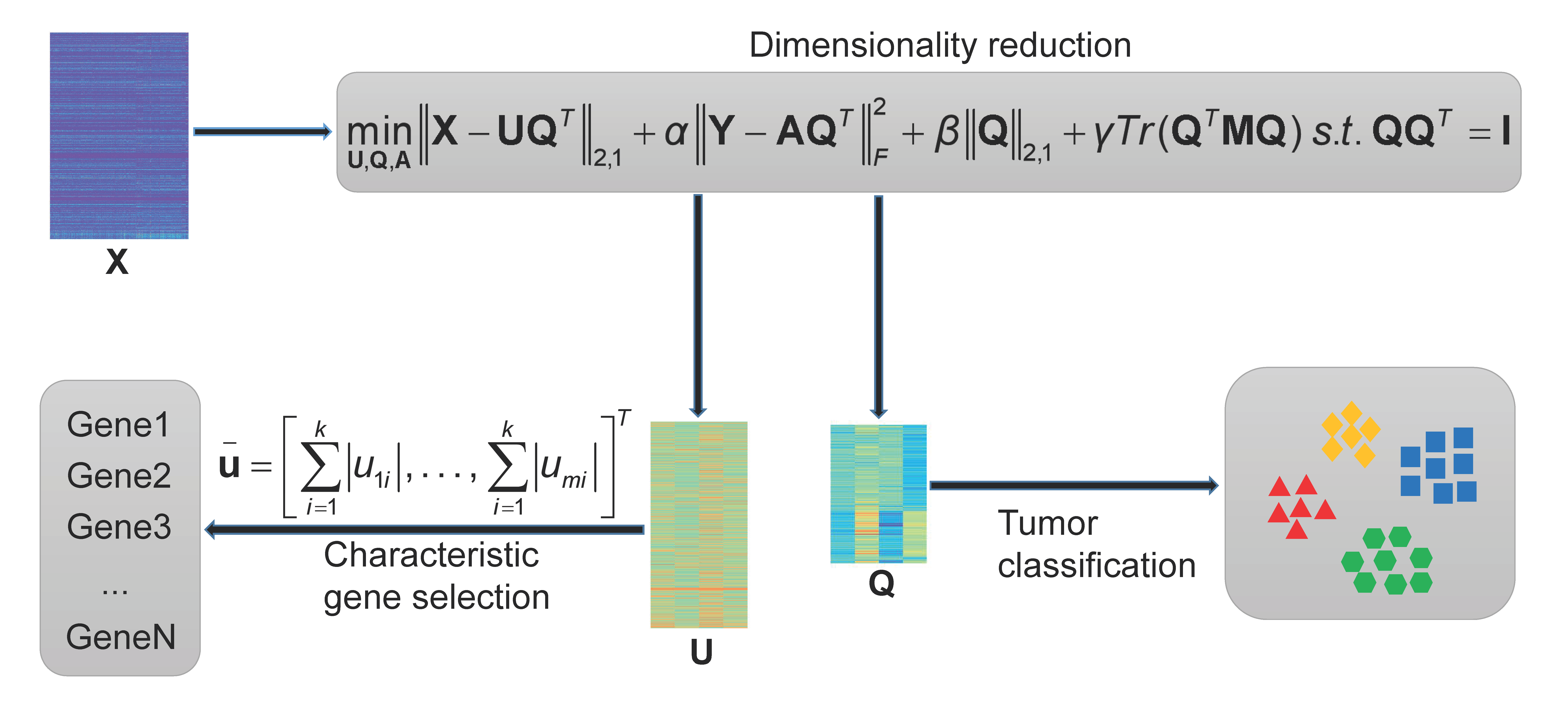 Framework of the proposed RLSDSPCA method.
Framework of the proposed RLSDSPCA method.